The CyanoTag Project
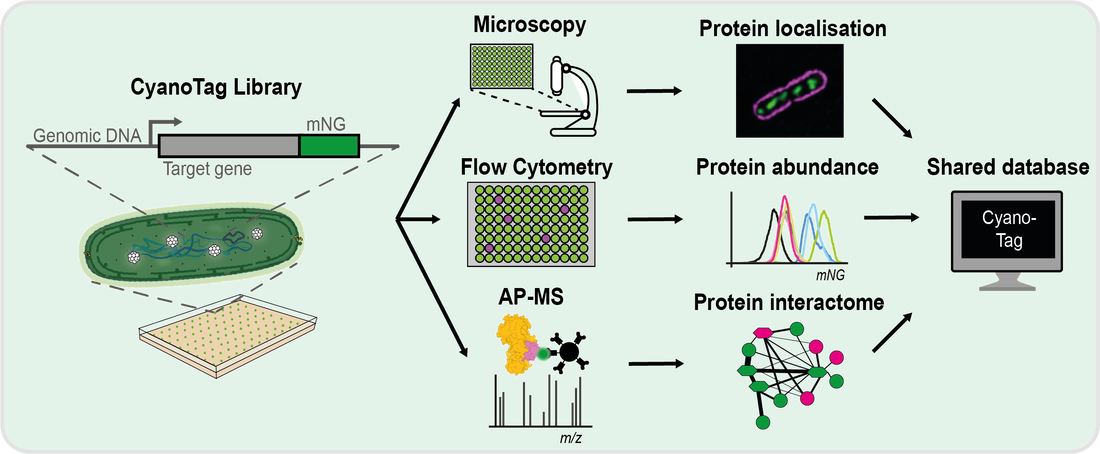
CyanoTag uses the power of high-throughput approaches and fluorescent tagging to discover more about the proteins of a model photosynthetic bacteria. Our pipeline for scarless endogenous fluorescent protein tagging in the model cyanobacterium Synechococcus elongatus PCC7942 allows us to determine subcellular localisation, track relative protein abundances and to elucidate protein-protein interaction networks. As of early 2024 we have targeted over a quarter of the 2714 proteins in S. elongatus. Associated imaging, expression and interactome data is publicly available and is shared via an online multi-omics database (MORF) and our recent preprint/publication. Detailed methods and additional resources will also be available to download from this page.
So far the CyanoTag project has provided insights into processes as diverse as photosynthesis and bacterial cell division, relevant to cyanobacteria and more broadly to all photosynthetic life. Our tagged lines, datasets and methods are intended to be shared, so as to facilitate future discoveries in these fascinating organisms.
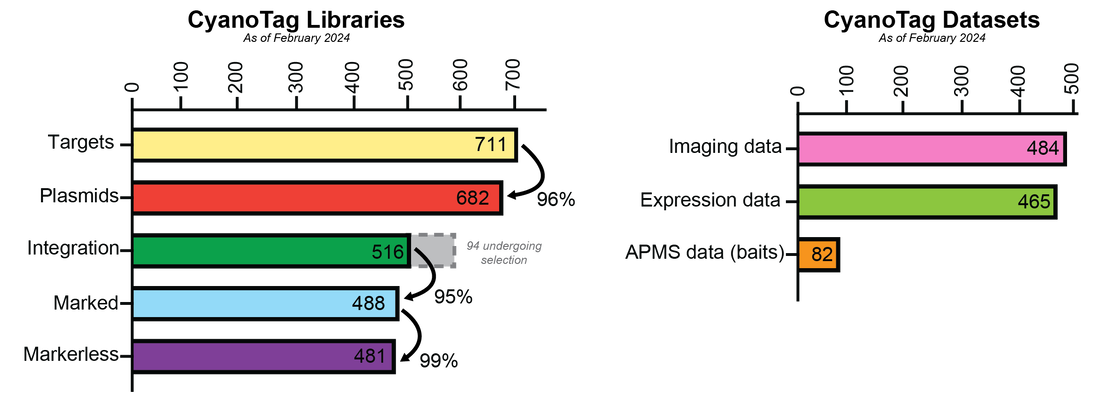
Current libraries and datasets
CyanoTag Project Highlights
....will be added here soon! In the meantime, check out our preprint.
Documents & Datasets
|
Methods document version 1.2 (April 2024)
Contains detailed, step-by-step methods for generating and working with tagged cyanobacteria Version 2.0 containing more detailed methods (including photographs) is in development. |
| ||||||
| |||||||||||||
|
Browse our data for over 700 protein targets in MORF via https://morf-db.org/projects/York-Mackinder-Lab/MORF000032 or via our latest data table.
|
| ||||||